Some Non-linear dimension reduction techniques (NLDR), such as
t-distributed stochastic neighbor embedding (tSNE) do not provide
mechanism to employ prediction like Uniform manifold approximation and
projection (UMAP). In that case, the predict_2d_embeddings
function serves as a valuable tool to predict 2D embeddings, regardless
of the NLDR technique.
model <- fit_high_d_model(training_data = s_curve_noise_training,
nldr_df_with_id = s_curve_noise_umap)
df_bin_centroids <- model$df_bin_centroids
df_bin <- model$df_binpredict_df <- predict_2d_embeddings(test_data = s_curve_noise_test,
df_bin_centroids = df_bin_centroids,
df_bin = df_bin, type_NLDR = "UMAP")
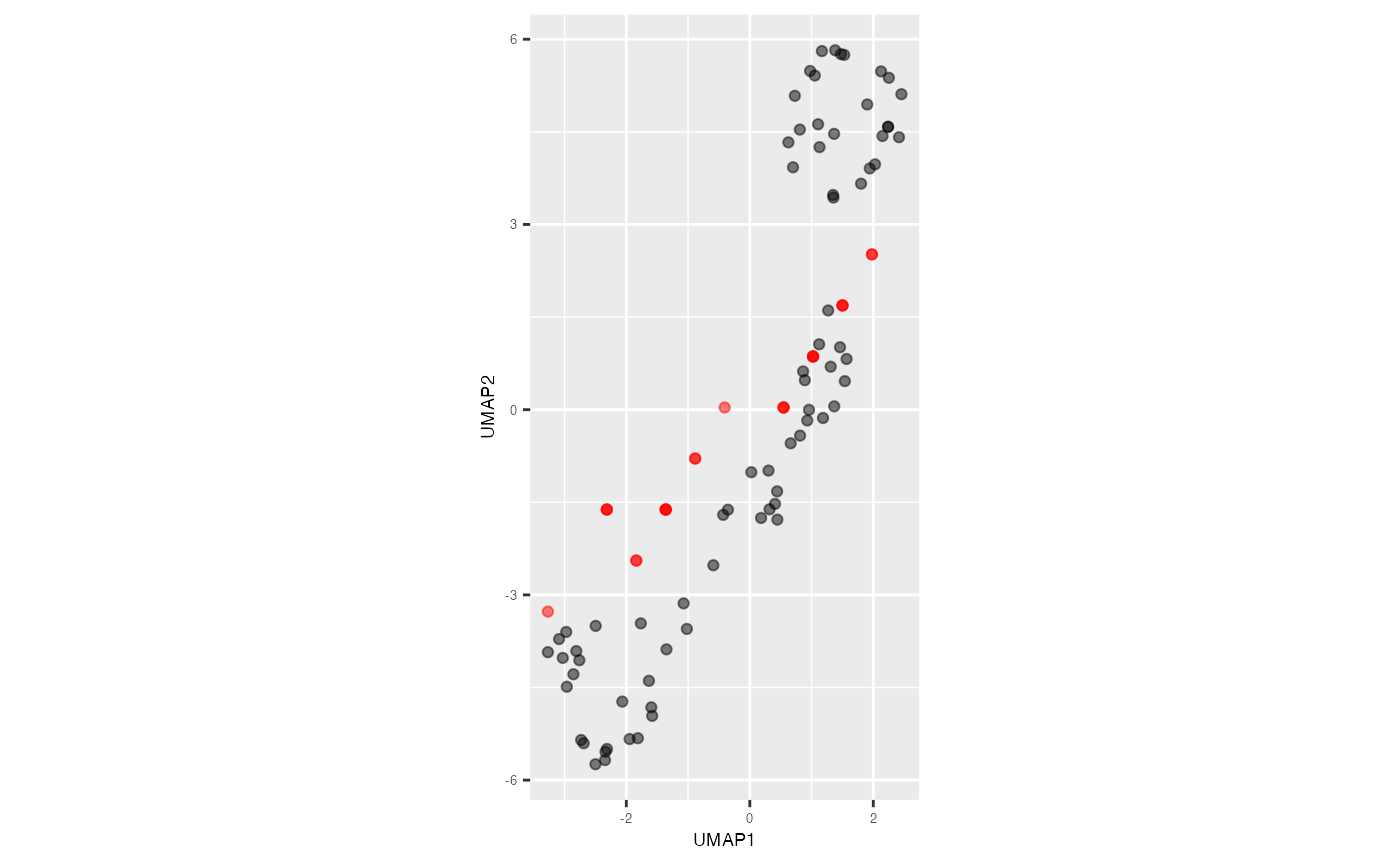
predict_df
#> # A tibble: 25 × 4
#> pred_UMAP_1 pred_UMAP_2 ID pred_hb_id
#> <dbl> <dbl> <dbl> <dbl>
#> 1 -1.84 -2.44 5 9
#> 2 1.02 0.862 10 40
#> 3 1.02 0.862 13 40
#> 4 -2.32 -1.62 18 16
#> 5 -1.36 -1.62 27 17
#> 6 -1.36 -1.62 28 17
#> 7 1.02 0.862 29 40
#> 8 1.50 1.69 30 48
#> 9 -1.84 -2.44 32 9
#> 10 0.547 0.0355 36 33
#> # ℹ 15 more rowss_curve_noise_umap |>
ggplot(aes(x = UMAP1,
y = UMAP2,
label = ID))+
geom_point(alpha=0.5) +
geom_point(data = predict_df, aes(x = pred_UMAP_1, y = pred_UMAP_2),
color = "red", alpha=0.5) +
coord_equal() +
theme(plot.title = element_text(hjust = 0.5, size = 18, face = "bold"),
axis.text = element_text(size = 5),
axis.title = element_text(size = 7))